DNA
It is estimated that approx. ten double-strand breaks (DSBs) occurs each day per cell that are based on breaks during metaphase in the chromosome and chromatid early passage primary human or mouse fibroblasts (Lieber et al. 2010). DSB in the DNA can be caused by both endogenous and exogenous factors such as exposure to irradiation such as ultraviolet light (UV) and chemical agents. The checkpoint mechanisms have been evolved that facilitate repair the DNA damage. Numerous repair pathways do exist which include base excision repair (BER), nucleotide excision repair (NER), and double-strand break repair (DSBR) (figure 1). The main repair pathways in DSBs are non-homologous end joining (C-NHEJ) and homologous recombination (HR) (Truong et al. 2014). DSBs are complicated as if not repaired properly can cause cell death. DSBs can lead to deletions, translocations, and fusions of DNA molecules (Negritto, 2010).
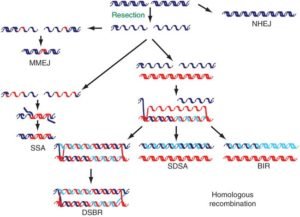
Figure 1: DNA double-strand breaks (DSBs) repairs mechanisms (Negritto, 2010)
In eukaryotes, double-strand DNA breaks are considered as common event. For the repair of DSBs two major pathways in eukaryotic cells are homologous recombination and nonhomologous DNA end joining (Lieber et al. 2010).
ZFNs, Domain structure of FokI endonuclease
FokI is a restriction enzyme that belongs to bacterial type IIS restriction endonuclease. The enzyme has the non-palindromic pentadeoxyribonucleotide recognition site i.e. 5′-GGATG-3′: 5′-CATCC-3′, in a duplex DNA thereby cleaving the 9 out of 13 nucleotides downstream of the recognition site. The enzyme does not have the ability to recognize any specific-sequence at cleavage site. The two distant protein domains are present within FokI having varied functions. One domain has sequence-specific recognition within the DNA whereas, other poses endonuclease activity. After the anchorage of DNA-binding domain at the recognition site, there is a transmission of signal to the endonuclease domain. The signal cause activation of the endonuclease activity by the allosteric interactions thus cleavage takes place. The domain structure of FokI restriction endonuclease revealed an N-terminal DNA-binding domain and a C-terminal domain with non-specific DNA cleavage activity. In of FokI endonuclease, single amino acid substitutions are linked to the decoupling DNA-binding as well as the associated strand scission activities. The crystal structures of FokI and FokI-bound to its related site, confirmed the FokI endonuclease modular nature in one of the study. The endonuclease domain is confiscated in a ‘piggyback’ manner by the recognition domain that have a protein–protein interactions. Endonuclease domain release from the recognition domain permits it to move towards the DNA cut site thus cleavage takes place. The cleavage occurs in Mg ion presence and binding of the FokI to its cognate site. Furthermore, FokI endonuclease domain dimerization is considered as key factor for the production of DSBs. The engineering of numerous novel chimeric endonucleases was generated by fusing the FokI cleavage domain with the further DNA-binding proteins in eukaryotic DNA-binding motifs. First chimeric endonuclease was generated by linkage of FokI cleavage domain with the Ubx homeodomain from the Drosophila (Durai et al. 2005).
Mechanism of DSBs generated by ZFNs
The Process of double-strand break by using ZFN necessitates nuclease domain to dimerization for cutting the DNA substrate. ZFN dimerization, The DSBs by ZFNs is facilitated by two meticulously oriented inverted binding sites having positioned in a head to tail orientation on the double stranded DNA substrate. The first evidence of FokI dimerization by use of its cleavage domain originated from the work on the FokI endonuclease crystal structure. Subsequently, kinetic studies support the view of dimerization by nuclease domain of FokI restriction enzyme and regarded as essential for the breakage of double-stranded DNA substrate.
Three-finger ZFN effectively has an 18 bp recognition site considered to be sufficient to specify a unique sequence in plants as well as mammalian genome. The above described mechanism for DNA cleavage showed delicate ZFNs sequence-specificity and application in different experiments in vivo for gene targeting in frog oocytes, Drosophila, plants and human cells.
TALENS and DSBs
In order to generate DSBs, TALE nucleases make use of a C-terminal fusion with the type II restriction enzyme FokI thus creating a heterodimer. The DSBs generated by the Nuclease activity of TALENs are repaired via nonhomologous end joining (NHEJ) or homologous directed repair (HDR). Introduction of the heterodimers for nuclease activity resulted in the minimization of off-target effects. In this process, binding of two TALEN proteins to the plus as well as minus strand of DNA thereby creating a FokI heterodimer ultimately generating a double-stranded break. The technique for studying the mutations, TALENs have been useful widely used to generate know down/ knock out genes by using nuclease activity in numerous organisms which include bacteria, yeast, plants, nematodes, fish, Xenopus embryos, human cell lines, rodents, and rat embryonic stem cells. TALE proteins applied to edit mammalian besides bacterial genome at specific locations within the DNA. This study showed that in E. coli longer targets of almost 26- 32 bp recombined 100-fold more proficiently compared to the 14 and 20 base pairs targets which are comparatively shorter. Similarly, when compared to mammalian cells ∼20 fold efficiency and ∼6 fold efficacy was observed with a 44 and 32 bp target, respectively (Moore et al. 2014; Joung and Sander, 2013).
Cas9 nucleases as DSBs machinery
Cas9 nuclease have been widely assumed in the field of genome-editing. Cas9 has the ability to site-specifically cleave double-stranded DNA, thereby generating the DSBs and ultimately activating the double strand break (DSB) repair machinery. Cas9D10A, one of the the Cas9 mutant is generated to increase the specificity and have only nickase activity with the ability to cleaves just one strand of DNA strand. Considering the target specificity of the Cas9D10A, targeting the locus with the paired Cas9 complexes intended to generate nearby DNA nicks. CRISPR/Cas9 system has been extensively accepted for targeting the genes in various organisms e.g. human, bacteria, zebrafish, C. elegans , plants, Xenopus tropicalis, yeast, Drosophila, monkeys, rats and mice. The application of CRISPR/Cas9 system for the introduction of single point mutations in a particular target gene due to the nuclease activity (Tool et al. Gilles et al. 2014).
ZFNs, TALENS, Cas9 nucleases in other organisms
The targeted mutagenesis and gene replacement in the Drosophila by using ZFN-mediated stimulation was initially specific to flies but later ZFNs are applied to other organisms. It was demonstrated that ZFNs are functional model plant, Arabidopsis thaliana as well. The experiments used different nucleases and ZFNs showed to be active in plants. Another experiment demonstrated the efficacy of ZFNs in tobacco cells with known specificity leading to homologous repair thereby regenerating whole plants. The nematode, C. elegans, is considered as an important model organism for biological studies in numerous fields like development, neurobiology and genetic studies. Studies utilised model organism to demonstrate the use of ZFNs for cleavage studies generating DSBs at different levels employing DNA as synthetic extrachromosomal target as well as an endogenous genomic target. The experiment using C. elegans are somewhat similar to the experiments with Drosophila genes. In these experiments there is an involvement of genetic engineering for the construction of novel sets of ZFs. However, the experiment by the use of ZFNs in C. elegans is not fruitful compared to the Drosophila. The advent of TALEN and CRISPR-Cas9 systems with nuclease activity showed modification in the genomes via mutagenesis in the worms. In another experiment that used Cas9 as nuclease in the C. elegans germline for the Ribonucleprotein delivery instead of vector DNA is known to be effectively in mammalian cells among other lower organisms (Chandrasegaran et al. 2016).
Three potential advantages that make TALENs effective when compared with the ZFNs are described. Above all others is the TAL domains modularity along with the simplicity in their DNA recognition code. This property allows TALENs to recognise and target almost any sequence within the genome of different organisms. Another aspect is that the specificity of TAL domains is not dependent upon the context as occurs in the ZFNs thus resultant is the prediction accuracy of target specificity with high rate of success. Finally, the experiments that make use of TALENS in the gene targeting in different organisms revealed efficacy of the TALENs yielding targeting efficiencies ranged between 30 to 100% in case of NHEJ-mediated knock-outs whereas 1-10% for HDR-mediated knock-in (Carroll et al. 2014).
DSBs formation during meiosis by SPO11
The initiation of meiotic recombination is favored by the complex of SPO11 i.e. the meiotic specific trans-esterase along with other accessory proteins that generates DSBs within the homologous chromosomes. The ability to form DSBs at specific sites known as hotspot regions are 100-1000 times greater compared to other regions. The accessory proteins including Rec114, Mer2, and Mei4 binds to axis site within chromosome followed by formation of a complex with DSBs thereby generating pre-DSBs recombinosome (figure 6, 7). For the dimerization of SPO11 accessory proteins involve include Rec102, Rec104, Ski8 allowing DNA binding and nuclear retention (Arora et al. 2004 and Panizza et al. 2011). Other Spo11 accessory proteins known as RMM are group of protein including Rec114, Mer2, and Mei4, forming sub-complex that partially localize on the chromatin and favor the binding of Spo11 to the DNA cleavage sites (Panizza et al. 2011).
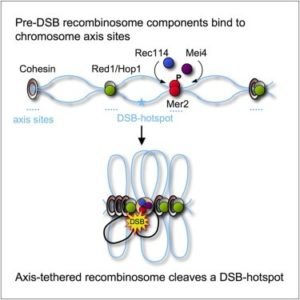
Figure 6: Pre-DSB recombinosome components
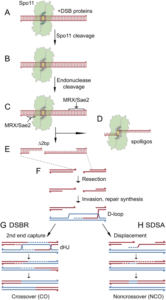
Figure 7: SPO11 dependent DSBs along with accessory proteins (Cole et al. 2010)
DSBs formation during meiosis by SPO11 in yeasts, C. elegans and plants
In Saccharomyces cerevisiae (yeast), meiotic recombination is a consequence of nine gene products which are meiosis specific. Among them six are well established i.e. SPO11, MEI4, MER2, REC102, REC104 and REC114 having role in the molecules formation and subsequent processing of DNA double strand breaks (DSBs) using specific pathway. Other proteins in yeast may be known to act as modulators in the DSBs formation in yeasts (Keeney et al. 1997). The DSBs and recombination events are thought to be conserved. Similarly, as in C. elegans DSB-generating enzyme is found to be homolog in yeast i.e. Spo11p, is required for meiotic exchange. In worms, primarily SC formation is thought to be a requisite for homologous synapsis is the stabilization of pairing rather than DSBs as in yeasts. The deletion of recombination promoting region do have implication in the defects in paring of chromosomes in C. elegans (Dernburg et al. 1998). In C. elegans, meiotic recombination is eliminated in absence of SPO11but SC are formed between the homologs normally. The single SPO11homolog is known to be present in animals and yeasts whereas three homologs exist in Arabidopsis thaliana as well as in higher plants. SPO11-1 and SPO11-2are known to act as the initiate meiotic recombination and SPO11-2 also ssary for proper chromosome segregation whereas SPO11-3 role towards somatic endoreduplication is reported (Hartung et al 2007).
References
- Hartung, F., Wurz-Wildersinn, R., Fuchs, J., Schubert, I., Suer, S. and Puchta, H., 2007. The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. The Plant Cell, 19(10), pp.3090-3099.
- Negritto, M. C., 2010. Repairing Double-Strand DNA Breaks. Nature Education 3(9), 26.
- Sundar Durai, Mala Mani, Karthikeyan Kandavelou, Joy Wu, Matthew H. Porteus, Srinivasan Chandrasegaran; Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells, Nucleic Acids Research, Volume 33, Issue 18, 1 January 2005, Pages 5978–5990, https://doi.org/10.1093/nar/gki912
- Cole, F., Keeney, S., & Jasin, M. (2010). Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes & Development, 24(12), 1201–1207. http://doi.org/10.1101/gad.1944710
- Moore, R., Chandrahas, A., & Bleris, L. (2014). Transcription Activator-like Effectors: A Toolkit for Synthetic Biology. ACS Synthetic Biology, 3(10), 708–716. http://doi.org/10.1021/sb400137b
- Gilles, A. F., & Averof, M. (2014). Functional genetics for all: engineered nucleases, CRISPR and the gene editing revolution. EvoDevo, 5, 43. http://doi.org/10.1186/2041-9139-5-43
- Tool, M., Calculator, T., Tool, P.S., Tool, N.G.G.A., Charts, S. and Guides, T., CRISPR/Cas9 & Targeted Genome Editing: New Era in Molecular Biology.
- Joung, J.K. and Sander, J.D., 2013. TALENs: a widely applicable technology for targeted genome editing. Nature reviews Molecular cell biology, 14(1), p.49.
- Chandrasegaran, S., & Carroll, D. (2016). Origins of Programmable Nucleases for Genome Engineering. Journal of Molecular Biology, 428(5 Pt B), 963–989. http://doi.org/10.1016/j.jmb.2015.10.014
- Carroll, D., 2014. Genome engineering with targetable nucleases. Annual review of biochemistry, 83, pp.409-439.

