Abstract
The identification of bacteria by morphology and other conventional methods are still used even though numerous high techniques are available. The recent study aims to explore conventional techniques for the identification of bacteria. Gram-staining can differentiate Gram-positive from Gram-negative bacteria and is regarded as the frequently used in-vitro technique for bacterial smears classification. The purpose of the recent study is to explore conventional techniques for identifying bacteria, ultimately leading to improvements in infectious disease diagnosis or identification of bacteria from environmental for industrial purposes. Gram staining, spore staining, capsule staining, and acid-fast staining techniques are employed for identification purposes. The results of different staining procedures showed that the sample might contain Bacillus species.
Introduction
Various techniques are present and used today for analyzing the microorganisms. These techniques may regulate more than one facet of a microbial system, including identification and characterization, and quantification. The techniques consist of differential staining, serological methods, protein analysis, and sequencing analysis (Desai and Armstrong, 2003). Rapid diagnosis for bacterial presence is vital for effective treatment of infected diseases as clinical signs are not enough to identify infectious agents. Although identifying microbes through traditional culture-based techniques is limited and time-consuming still are in use worldwide (Sandra et al. 2016). The identification of bacteria by morphology and staining methods is considered conventional but still used in microbiology. Modern techniques for identification are available, such as genome typing to identify and detect novel microbes and the determination of previously uncharacterized microorganisms (Fredericks and Relman, 1996). The recent study aims to explore conventional staining techniques for the identification of bacteria. However, staining techniques are considered to be cost-effective compared to the recent genome-based techniques. Moreover, the Gram staining can differentiate Gram-positive from Gram-negative bacteria and is regarded as the frequently used in-vitro technique for bacterial smears classification.
Materials
Tryptic soy agar slants
Gram stain kit
Spore stain kit
Acid-fast stain kit
capsule stain kit
Methods
Obtain the sample containing unknown bacteria from the broth culture or slant. Then perform gram staining procedure. Add primary dye; after few minutes, the decolorizing agent is added, followed by washing and added counterstain. Examine under a microscope. For endospore staining, use sporulating media or further incubate the culture for additional 24-48 hrs and then perform spore staining. Use 24 hrs tryptic soya agar slants culture label with gram stain as a positive control. Similarly, use acid-fast positive bacterial culture stain as a positive control for acid-fast staining procedure.
Gram Staining
The microbial cell wall staining technique was employed using crystal violet as the primary dye for Gram differentiation (Lamanna and Mallette, 1954).
Acid Fast staining
Ziehl-Neelsen method was used for the identification of acid-fast bacteria (Putt, 1951).
Spore Staining
Schaeffer spore staining method was used for endospore staining (Ashby, 1938).
Capsule staining
The wet-mount method was used for capsular staining. India ink is the staining dye in which the organism possessing a capsule is visualized as a refractile zone surrounding a cell (Baker, 1920).
Results
Figure 1 shows that the morphology of bacteria in the sample are rod-shaped present in clusters. Gram staining results showed gram-positive rods as stained purple in the figure.
Figure 2 showed acid-fast staining intense purple in the form of straight or slightly curved under the microscope. The bacteria that appeared red after staining are positive for the presence of acid and alcohol fast bacillus. The microbes may exist in groups (A) or single-cell (B). thus, are negative results with acid-fast staining.
Figure 3 depicted capsular staining. The results after staining showed that unknown bacteria in the sample do not have the ability to form capsules.
Figure 4 illustrates the result of spore staining. As shown in the figure, vegetative cells are stained pink (A). At the same time, endospores are stained green and visualized as ellipses.
Accordingly, the results of different staining methods used for the unknown bacteria in the sample are gram-positive rod, clustered, or present as a single cell. Non-capsular, spore-forming, and showed a negative result for the acid-fast staining.
Gram staining results showed the possibility of Bacillus subtilis, Corynebacterium xerosis, and mycobacterium smegmatis. Further, spore staining ruled out the possibility of Corynebacterium xerosis and Mycobacterium smegmatis as both are non-spore-forming bacteria. Thus, the interpretation of the staining results identified Bacillus subtilis in the unknown sample.
Discussion
The Gram stain reports were highly accurate and remained so over the years studied. The performance characteristics for the main morphological groups were close to 100%. Figure 1 showed both grams positive and gram-negative rods, i.e., purple and pink colored rods, respectively. The appearance of gram-negative may be due to the comparatively low sensitivity for gram-positive rods towards gram staining. The low tendency of Bacillus spp. was also reported in the previous study (Søgaard et al., 2007).
To aid the results obtained via gram stains, wet mounts were employed and previously showed to provide help in interpreting Gram stains. The wet mount mostly cleared the ambiguity in gram-positive cocci arrangement and weakly stained gram-negative bacterial cultures appearing more discrete in wet mounts (Søgaard et al. 2007).
The spore-forming gram-positive or gram-variable rods showed that either Clostridium or Bacillus species are likely to be present in the sample under test. Even though Clostridium species are originally gram-positive bacilli that may or may not contain spores, the previous finding reported that they often appear gram-negative by staining in clinical specimens of Clostridium organism (Barenfanger and Drake, 2001). The results further confirm that the unknown sample belongs to the Bacillus.
Modifications in the conventional staining techniques can improve and differentiation among bacterial species. The ultimate aim is to provide a cost-effective, improved method for identification, ultimately leading to advancements in the clinical application towards a better diagnosis of infectious agents.
References
- Fredericks, D.N. and Relman, D.A., 1996. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clinical microbiology reviews, 9(1), pp.18-33.
- Desai, M. J., & Armstrong, D. W. (2003). Separation, identification, and characterization of microorganisms by capillary electrophoresis. Microbiology and molecular biology reviews, 67(1), 38-51.
- Becerra, S. C., Roy, D. C., Sanchez, C. J., Christy, R. J., & Burmeister, D. M. (2016). An optimized staining technique for the detection of Gram-positive and Gram-negative bacteria within the tissue. BMC research notes, 9(1), 216.
- Lamanna, C. And Mallette, M. F. 1954 The cytological basis for the role of the primary dye in the gram stain. J. Bacteriol., 68, 509-513.
- PUTT, F. A. (1951). A modified Ziehl-Neelsen method for demonstration of leprosy bacilli and other acid-fast organisms. American journal of clinical pathology, 21(1), 92.
- Ashby, G. K. (1938). Simplified Schaeffer spore stain. Science, 87(2263), 443-443.
- Baker, S. L. 1920. Technique for the demonstration of the capsules of bacteria. Br. J. Exp. Pathol. 1:127.
- Søgaard, M., Nørgaard, M., & Schønheyder, H. C. (2007). First notification of positive blood cultures and the high accuracy of the gram stain report. Journal of clinical microbiology, 45(4), 1113-1117.
- Barenfanger, J., & Drake, C. A. (2001). Interpretation of Gram stains for the non microbiologist. Laboratory medicine, 32(7), 368-375.
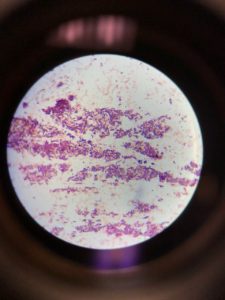
Figure 1: Gram staining (A). Gram-positive rods in the cluster; (B). Single-cell Gram-positive rod; (C). Gram-negative rods appeared as pink
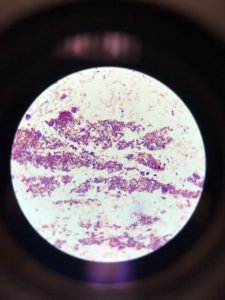
Figure 2: Acid Fast staining arrowhead (orange) represented clustered dark purple stained straight or slightly curved rods
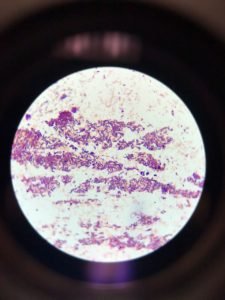
Figure 3: Capsular staining illustrated the absence of hollows around the bacterial cells
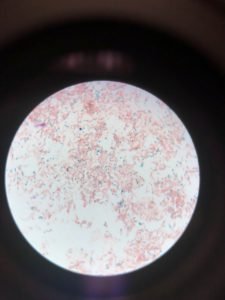
Figure 4: Spore Staining (A). Vegetative cells stained pink; (B). Endospore

